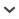Since first human settlement c. 400 years ago, the Mascarene islands have undergone some of the highest rates of ecosystem transformation and species extinction recorded worldwide. One surviving species, the Mascarene Swiftlet Aerodramus francicus native to the islands of Mauritius and Réunion, is typical among Aerodramus swiftlets in presenting a paucity of distinguishable morphological characters, as well as being particularly vulnerable to anthropogenic activities. In this study, a primary objective was generating genetic data for each island population, allowing genetic divergence between them to be assessed for the first time, employing informative methods regarding the taxonomic status. We find that the two island populations are 0.8% divergent and reciprocally monophyletic in the mitochondrial ND2 gene, with nuclear data (β-fibrinogen intron 7) being largely congruent but showing incomplete lineage sorting and low resolution. Results of barcode gap and coalescent species delimitation analyses are consistent with a comparison of the genetic divergence uncovered within Mascarene Aerodramus with that among other long-established Aerodramus species. The findings of all three approaches are consistent with the two island populations representing two separate species. However, because of lack of resolution in the nuclear locus, we conservatively retain a single species pending future data. To assess the conservation status, we conducted the first quantitative assessment of population size and distribution through extensive nest counts on the islands of Mauritius and Réunion, and by considering the threats to swiftlets on each island. The survey produced estimates of 10,100–10,700 individuals for Mauritius, and 39,600–53,500 individuals for Réunion. Considering the population size data, inferred distributions, and numerous conservation concerns, an updated conservation status to Threatened (Endangered): B2 b(i,ii,iii,iv,v) c(i,ii,iii,iv) is warranted according to IUCN guidelines. Improving protection of the breeding habitat of the Mascarene Swiftlet is crucial to prevent this endemic species from being added to the Mascarene's staggering extinction toll.
The biotas of oceanic islands are known to be particularly vulnerable to the effects of human activity, including habitat loss and the introduction of exotic species (Gillespie & Clague 2009). As a result of both this high intrinsic vulnerability of island populations, and high levels of endemism in island archipelagos, island species represent a disproportionately high percentage of global biodiversity under threat from anthropogenic change (Myers et al. 2000).
The islands of the Mascarene archipelago (Réunion, Mauritius and Rodrigues), Indian Ocean, are a case in point. Since first arrival in the archipelago c. 400 years ago, humans have directly or indirectly caused the extinction of 50–60% of the native vertebrate fauna, including birds, bats, giant tortoises and other reptiles (Cheke & Hume 2008, Probst & Brial 2002). Given the relatively high level of economic development of the Mascarenes compared with other archipelagos worldwide, it might be expected that the remaining fauna would be well protected. Indeed, the Mascarenes is home to major conservation programmes, including some internationally recognised conservation success stories (Jones et al. 1995). However, one of the islands (Mauritius) has also been home to deliberate anti-conservation choices (Florens 2012a,b, 2013). Furthermore, conservation efforts have necessarily been focussed on the most threatened (and often the most emblematic) species, while other populations have received less attention, even among vertebrates. Such is the situation for the Mascarene Swiftlet Aerodramus francicus, endemic to the islands of Mauritius and Réunion, which nests and roosts in caves in colonies that may attain many hundreds of pairs (Middleton 1998b). Since Mascarene Swiftlets must return to the colony at night, they are extremely sensitive to a wide range of anthropogenic pressures, which include destruction of colonies due to peri-urban expansion (Hammond et al. 2015), cave tourism, illegal rubbish dumping and the poaching of nests for bird's nest soup. A further threat comes from the use of an exceptionally high quantity of pesticides per area of cropland on Mauritius (FAO 2022). As a consequence of such threats, swiftlet numbers have undergone major declines on both islands. Over the last 2–3 decades, the most extreme destruction and decline of colonies is thought to have taken place on Mauritius (Cheke & Hume 2008, Safford & Hawkins 2013). Currently the species is classified by the IUCN as Near Threatened (BirdLife International 2022). However, this considers all Mascarene Swiftlets as a single taxon and does not take into account differences in threats and demographic status between Mauritius and Réunion.
Mascarene Swiftlets are typical of insular Aerodramus swiftlets at a global scale, not only being of conservation concern, but also presenting challenges for taxonomy due to a paucity of obviously distinguishable morphological characteristics, in particular plumage variation. Mayr's (1937) summary for the Indo-Australian region is equally true for Indian Ocean islands: “every author who has ever worked with these small swiftlets (...) will contend that their classification presents the most difficult problem in the taxonomy of birds”. Recently, it has been shown that the Mauritius and Réunion populations of the Mascarene Swiftlet show phenotypic differences, in both plumage and biometrics (Kirwan et al. 2018), meriting at least subspecific status. Accordingly, the Réunion population was named A. francicus saffordi and the Mauritius population A. francicus francicus. However, there has currently been no attempt to compare genetic data for the Mauritius and Réunion populations. Among alternative datasets employed in systematics, genetic data have multiple benefits. First, despite limitations, the molecular clock has proven a valuable concept for inferring divergence times (Bromham et al. 2018). Compared with many phenotypic characters, including avian plumage, there exist multiple genes that evolve at a relatively regular rate, including those that are informative for molecular phylogenetics. Furthermore, genetic data are an important source of information on degrees of reproductive isolation between populations which underpins the biological species concept (Campillo et al. 2020).
During the last 15 years, many methods have been developed that employ both information on genetic divergence and hiatuses in gene exchange between populations to make explicit inferences of species status. Such species delimitation methods can be grouped into two broad categories. The first category corresponds to exploratory methods which propose partitions of species using the principle of genetic distance or more specifically the barcode gap, namely that for any single universally applied genetic marker, interspecies variation should be greater than the intraspecies variation (Meyer & Paulay 2005, Puillandre et al. 2021). The second category groups together methods based on coalescent theory that use multi-locus data to infer genetically isolated populations (Fujita et al. 2012, Rannala et al. 2020). In particular, the multispecies coalescent model (MSC) describes the probability distribution of the gene tree underlying a sample of DNA sequences from two or more genetically isolated populations (Rannala et al. 2020). The latter group of methods of species delimitation therefore has the advantage that it is not only repeatable by independent researchers, but is also among the few that attempt to infer evolutionary independence in terms of reduced gene flow.
This study has three main objectives. First, we obtain mitochondrial and nuclear data from both the Mauritius and Réunion populations of the Mascarene Swiftlet, with the objective of assessing the degree of genetic divergence and likely taxonomic status of the populations of each island. Second, we generate and assemble the most recent and complete colony count data to obtain the first justified estimate of the population size of each island. Lastly, we advance broad-scale conservation priorities for Mascarene Aerodramus in light of our findings on taxonomic status, population distribution, size and size trends, as well as knowledge of current threats.
METHODS
Sampling
We obtained a total sample of 19 individuals of the Mascarene Swiftlet. In addition to published sequences of two individuals from Mauritius (Johnson & Clayton 1999, Price et al. 2004), we sampled a further seven individuals from Mauritius and ten from Réunion. Tissue samples from Réunion were obtained from the bird rehabilitation centre of the Société d'Etudes Ornithologiques de La Réunion (SEOR) under Museum National d'Histoire Naturelle ministerial derogation, while blood samples from Mauritius were obtained through mist-netting and blood sampling with permits from the National Parks and Conservation Service, Mauritius. Samples were stored in ethanol and Queen's lysis buffer (Seutin et al. 1991).
Sequences were generated for the mitochondrial gene encoding the NADH dehydrogenase protein subunit 2 (ND2), intron 7 of the nuclear gene β-fibrinogen (Fib7) and one sequence for the mitochondrial gene encoding for cytochrome b (Cytb). PCR amplifications and sequencing were performed using primers L5215 (Hackett 1996) and H6313 (Johnson & Sorenson 1998) for ND2, L14841 (Kocher et al. 1989) and H4a (Harshman 1996) for Cytb and FIB-B17U and FIBB17L for Fib7 (Prychitko & Moore 1997) and were carried out in a volume of 25 µL including 2 µL of solution containing the DNA extraction, 1.25 µL for the two primers and 0.125 µL of Qiagen Taq. We then used thermal cycling procedures comprising an initial denaturation of 2 min at 94°C followed by 35 cycles of 30 s at 95°C, 30 s of hybridization temperature: 54.2°C and 30 s at 72°C for elongation. Sanger sequencing was performed by Eurofins Genomics. Sequences were then checked and cleaned using the chromatograms provided by Eurofins. Extra published Aerodramus sequences and those of other members of Apodidae (outgroup) were downloaded from GenBank and supplied by the authors (Johnson & Clayton 1999, Price et al. 2004). The selected outgroup species are Alpine Swift Apus melba and Pygmy Swiftlet Collocalia troglodytes. These species were chosen as an outgroup because they are relatively close to the group of interest, Indian Ocean Aerodramus (ingroup), while being distant enough to help establish the monophyly of this group (Price et al. 2004). The sequences were then assembled and edited using Geneious v. 5.4.7 (using Geneious Alignment, cost matrix: 65%, gap open penalty: 12, gap extension penalty: 3). Individuals with missing data for ND2 or Fib7 were pruned from alignments used for phylogenetic and species delimitation analyses, but retained for analyses of genetic distance.
Phylogenetic analyses
The phylogenies for all analyses were estimated using maximum-likelihood (ML) as implemented in RAxMLHPC2 v. 8.2.12 (Stamatakis 2014) and a Bayesian inference approach performed using BEAST2 v. 2.6.6 (Bouckaert et al. 2019) using the computational resources of the CIPRES Science Gateway (Miller et al. 2010). We performed Bayesian inference phylogenetic analysis on a concatenated ND2 and Fib7 matrix and the ND2 single gene matrix, and Maximum Likelihood analyses of both the concatenated matrix and the separate Fib7 and ND2 single gene matrices. The nucleotide substitution models best fitting the data were computed for each gene matrix using a hierarchy implemented in the software jModelTest2 v. 2.6.6, and the best model selected using the Bayesian information criterion (Darriba et al. 2012). The substitution models were applied in BEAUti v. 2.6.7 to each corresponding gene in the matrices (Bouckaert et al. 2019). Parameter values of nucleotide frequencies and (depending on the type of model) conversion rates, proportion of invariable sites, and gamma distribution were estimated by the BEAST program.
A relaxed log normal molecular clock was applied with clock rates partitioned for each gene from previously estimated evolutionary substitution rates of nuclear and mitochondrial genes (Lerner et al. 2011). Confidence intervals for divergence times were automatically computed in BEAST. BEAST phylogenies were reconstructed using MCMC for Bayesian inference. Two independent chains of 10 million generations were run with trees sampled at every 1000th generation for a total of 20,000 trees. The convergence of the chains was assessed using Tracer v. 1.7.1. LogCombiner was used to combine the two runs after discarding the first 10% of trees (burn-in period), and TreeAnnotator was used to produce a maximum clade credibility tree with mean node heights. For ML analyses we used the rapid bootstrapping algorithm available in RAxML. Furthermore, we calculated pairwise HKY genetic distances between Aerodramus species in R v. 4.2.1 (programming script and distance matrix are available on GitHub:
https://github.com/RoriWijnhorst/MascareneSwiftlets GeneticDistance; R Core Team 2022).
Species delimitation
We first used the multispecies coalescent model (MSC) for species delimitation employing the combined ND2-Fib7 dataset as implemented in the Bayesian Phylogenetics and Phylogeography (BPP) program (Yang 2015, Flouri et al. 2020). We evaluated a scenario in which the Mauritius and Réunion populations of the Mascarene Swiftlet are candidate species, along with the ten other previously recognised species in our phylogeny. This method evaluates support for alternative hypotheses of species delimitation and species phylogeny, while accommodating conflicts between gene-trees and species-trees (Yang & Rannala 2010, 2014). Since prior distributions on the ancestral population size (θ) and root age (τ) can affect the posterior probabilities for models (Yang & Rannala 2010), we first performed a preliminary analysis estimating the two parameters under the MSC model with a given phylogeny (A00 configuration, using the topology obtained with the concatenated ML analysis). We then jointly estimated species delimitation and the species tree (A11 configuration) using both lower and higher estimates of inverse-gamma prior distributions of θ and τ as indicated by the A00 analysis – IGθ (3, 0.002), IGτ (3, 0.001), and IGθ (3, 0.003), IGτ (3, 0.04), respectively. We allowed rate heterogeneity across loci and used a heredity scalar of 1 for nuclear DNA and 0.25 for mitochondrial DNA. Each BPP analysis was run twice to check consistency of results between runs. Runs were conducted for 500,000 generations, with the first 10% of generations discarded as burn-in.
As an alternative to the multispecies coalescent model, we compare results with those obtained when employing the principle of the barcode gap, namely that for any single universally applied genetic marker, interspecies variation should be greater than the intraspecies variation (Meyer & Paulay 2005). In this context, we used the program Assemble Species by Automatic Partitioning (ASAP), which automates identification of the barcode gap. The algorithm first identifies gaps in the distribution of pairwise genetic distances and uses it to partition the dataset. Gap detection is then applied recursively to identified partitions, giving rise to finer partitions, until no further partitions are identified. Alternative partitions are ranked with a scoring system, computing P-values that any identified group forms part of a panmictic species under a neutral coalescent model (Puillandre et al. 2020). We performed three analyses for ND2 and Fib7 separately for each of the usable models: JC69, K80 and Simple Distance. While this method has the limitation that it can only be applied to any one single locus at a time, it has the advantage that it takes an agnostic approach to species delimitation, in that no specification of candidate species is required.
Nest counts and population abundance
Due to the behaviour of the Mascarene Swiftlet, which nests in colonies, and for which presence and numbers counted at any one moment are strongly associated with specific weather conditions and insect activity, distance-sampling techniques were considered inappropriate for estimating overall abundance. Furthermore, relative to the situation for continental populations, the islands of Mauritius and Réunion are relatively small and it is therefore feasible to count nests to estimate population sizes. Such counting is facilitated by the fact that swiftlets in the Mascarenes appear to nest year-round, with active nest building documented all twelve months of the year (Cheke 1987). Regarding Mauritius, we believe that it is possible to count all the colonies since the island has a flat topography and thanks to Middleton's work in the 1990s the island's cave system has been well explored and documented (Middleton 1998a,b). Réunion has a much more extreme topography due to rapid erosion following relatively recent volcanic activity forming multiple high mountains, one exceeding 3000 m. As a result of this extreme relief, many potential nesting sites are inaccessible. In addition, based on the known occurrence of colonies it appears impossible to characterise sites suitable for swiftlets from data readily available such as depth, or width of the entrance. A preliminary attempt to randomly sample caves was time-consuming and relatively uninformative, since all eight randomly-selected caves had no swiftlet nests. Thus, our estimate for Réunion is instead based on both systematic and random counting of all colonies documented in the literature covering the last 150 years and including 20 years of intensive observation conducted by the SEOR and other contributors.
We developed a protocol to best estimate the number of nests, including both the systematic counting and random components of our sampling. For systematic counting on both islands, we generated ranges in all cases in which it was difficult to be certain what constitutes one nest or multiple nests, with minima and maxima corresponding to the most conservative versus liberal count of whole nests. Remains of nests that were not whole were not counted. For Mauritius, the flatter terrain meant that all colonies could be systematically counted, and this was carried out between April 2019 and August 2021. The situation on Réunion is different since the difficulty of the terrain makes accessing many colonies not only arduous and time consuming, but also too perilous to envisage systematic counting in less than ten years with available funds. Documented locations of some colonies come from records that are up to 150 years old and usually do not include the number of nests. Since the 1990s, the SEOR has recorded all colonies reported in the literature and in the ‘faune-reunion’ citizen science database, while archiving their own counts made throughout Réunion since their foundation in 1997. Ten colonies, including what are unambiguously the three largest (La Chapelle, La Porte and the tunnel of the disused railway at La Possession) were counted between November 2015 and June 2021, while random sampling during May–July 2021 was used to estimate the number of nests in all other colonies. Specifically, using the subset of the other colonies that are accessible and a random-number generator (in R), a random selection was made of five different locations presented in the literature, whose nest counts were used to estimate the size of all other colonies that could not be counted. The size of the Réunion population was then estimated by combining the count of colonies systematically counted, with an estimate of the number of nests in the colonies that have not been systematically counted, based on the five locations randomly sampled and applying an adapted formula from Bibby et al. (2000) for population estimates from randomised surveys (Methods S1).
The size of the breeding population of Mauritius and Réunion was estimated by multiplying the number of nests by two. This method is commonly used (Tarburton & Tarburton 2013, Manchi & Sankaran 2014) even though it has certain limitations (Johnson et al. 2018). Here, we make the assumption that a specific number of birds, two in the case of A. francicus, are associated with one nest. This is a reasonable assumption because Aerodramus are monogamous and both parents care for their offspring (Koon & Cranbrook 2002, Safford & Hawkins 2013). However, to determine the effective population size (Ne) containing all mature individuals, we need to account for nonbreeders, i.e. individuals that were not nesting during our surveys but may breed at a later or earlier time. Since, we do not have information on the number of adult nonbreeding individuals, we used best available evidence for the ratio of breeding to nonbreeding individuals. For other Aerodramus swiftlets, it was estimated that nonbreeders may constitute c. 32% of the total adult population in Aerodramus maximus and 30–33% in Aerodramus fuciphagus (Nguyen Quang et al. 2002, Rahman et al. 2018). Using the highest and lowest estimates, we calculate a lower and upper bound for the total population size using the following equation:
Here, the breeding population is thus a maximum of 70% and a minimum of 67% of the total population. For the functional estimate of the total adult population, we use the mean of the lower and upper bound estimates.
Assessment of conservation status
We assessed conservation status against IUCN criteria (IUCN Standards and Petitions Committee 2022) applying our population size estimates. We also used the GPS locations of the colonies to calculate the extent of occurrence (EOO) and area of occupancy (AOO) using the freely available GeoCat tool (Bachman et al. 2011). The EOO is defined as the geographic range size of the species and computed by calculating the area of the smallest polygon that contains all sites of occurrence. The AOO is defined as the area in which the species occurs. This is automatically computed by summation of the area of square grids (at the recommended 2 km × 2 km scale) that the species occupies (IUCN Standards and Petitions Committee 2022). Additionally, we investigated which threats jeopardise the existence of the Mascarene Swiftlet. In addition to our collective experience of threats to Mascarene Swiftlets spanning 40 years, we consulted literature sources and other local experts.
RESULTS
Phylogenetic analyses
We obtained sequences of 1040 base pairs for the ND2 gene and 817 base pairs for Fib7.
JModeltest2 identified the TrN + I model of DNA substitution as best describing the data under the Bayesian Information Criterion for ND2, while it identified the TPM2uf model for Fib7. The closest hierarchically encompassing model was used in BEAST and ML analyses. In both ML and Bayesian analyses of ND2, the Mascarene Swiftlet populations of Mauritius and Réunion are divergent (0.8% pairwise HKY distance) and reciprocally monophyletic, with all nodes being well supported (PP = 1; bootstrap support (BS) = 90; Figure 1 and S1). The dated Bayesian tree yields a divergence of 0.1 million years between the Mauritius and Réunion populations (Figure S1).
The ML Fib7 tree has few nodes with strong support values, and the few supported nodes (BS ≥ 70%) are congruent with the ND2 trees. Indeed, a partition homogeneity test (Farris et al. 1995) on the combined ND2-Fib7 data (2 partitions, 1857 bp) indicated that the signal in the two gene regions does not differ significantly (P = 0.78). Furthermore, regarding our focal taxa, the Mascarene Swiftlet is supported as monophyletic (70% BBS), as is the Réunion-endemic lineage (86% BS; Figure S4).
The concatenated ND2-Fib7 ML and Bayesian trees are largely congruent with the ND2 trees. Specifically, in the case of the Mascarene Swiftlet, although the Mauritian population is not monophyletic in the concatenated ND2-Fib7 trees (Figure S2, S3) both the Réunion population and the Mascarene Swiftlet as a whole are monophyletic with strong support (PP = 0.81, BS = 50, and PP = 1, BS = 100, respectively). In both the concatenated ND2-Fib7 and ND2 tree, a sister relationship between A. francicus and Seychelles Swiftlet A. elaphrus is well supported (PP = 1; BS = 100; Figure 1 and Figure S1, S2, S3).
Figure 1.
Maximum likelihood analysis of the ND2 gene matrix based on GTR model for Aerodramus species and outgroups (Apus, Collocalia). Maximum likelihood bootstrap support values (1000 replicates) are indicated on nodes. Outgroup species were cropped from the figure to save space.

We further calculated pairwise genetic distances (Table 1). The genetic distance between Mascarene Swiftlets from Réunion and Mauritius is 0.8% for ND2 and 1.0% for Cytb. This is comparable to other relations between long-established Aerodramus species (Cibois et al. 2018, Price et al. 2004, Figure 1, S1, S2, S3) within the Aerodramus genus, such as the divergence between Seychelles Swiftlet and Mascarene Swiftlet (0.7% for ND2 and 1.2% for Cytb) and between Atiu Swiftlet A. sawtelli and Mariana Swiftlet A. bartschi (1.0% for ND2 and 0.8% for Cytb).
Species delimitation
Our BPP analyses for ND2-Fib7 yield consistent results, both with different prior distributions on the ancestral population size (θ) and root age (τ), and from independent runs with the same distributions. The Mauritius and Réunion lineages of Mascarene Swiftlet are consistently recovered as separate species (PP = 1.0), as are the ten previously recognised species in the dataset (PP > 0.97 in all cases). Results for our ASAP analysis for ND2 concord with those of the BPP analyses. Regardless of the distance metric employed (JC, K80 or Simple Distance), the partition with the highest ASAP score recovers the Mauritius and Réunion lineages as separate species, along with each of the ten previously recognised species. However, it should be noted that the ASAP analysis of Fib7 does not allow delineation of species reliably since with all three distance metrics, a minimum of three pre-established species are grouped together as one species (A. elaphrus, A. brevirostris and A. sororum, regardless of the distance metric used). Furthermore, there exist two partitions of equal ASAP-score, one of which recovers A. francicus as a single species, while the other groups A. francicus, A. elaphrus, A. brevirostris and A. sororum as a single species. These results are consistent with the lack of resolution observed in Fib7 in our phylogenetic analyses.
Figure 2.
Locations of colonies of A. f. saffordi on Réunion and A. f. francicus on Mauritius. The EOO of the two subspecies is shown by the area in dark grey and the EOO of the A. francicus as a whole. The EOO is the area of the smallest polygon containing all sites of occurrence. The AOO is calculated by summation of the area of red squares (2 × 2 km) in which colonies are found.

Table 1.
Summary of pairwise HKY genetic distances within the Aerodramus genus for mitochondrial genes ND2 and Cytb. Divergence between Mauritius and Réunion populations of A. francicus (underlined) can be compared with that among Aerodramus species from outside the Mascarenes.

Nest counts
On Mauritius, 103 caves were surveyed of which 37 contained swiftlet nests. Based on these surveys, we counted a total of 3514 nests (range: 3505–3537 nests). In order to safeguard the locations and breeding habitats of the swiftlets, the full data is only accessible to researchers upon request. The nest count data, where locations are masked, are available in the GBIF database ( https://doi.org/10.15468/k33fka). The two largest colonies documented by Middleton (1998b, 2000) for the 1990s (Surinam and Palma Caves, then estimated at 600–700 swiftlets and 350+ swiftlets, respectively) have since been lost, with no nests remaining in either cave in January 2020. An increase in numbers has however occurred in two of Middleton's (1998b, 2000) colonies (names withheld), the larger of which has increased in size tenfold to 1387 nests. Of the 34 colonies documented by Middleton (1998b, 2000) for the 1990s, there were only two small colonies that we were unable to survey (Quinze Cantons, 10–30 nests, Middleton pers. comm, access not granted by landowner; Chicken Manure Hole, 10 nests, not able to locate from available information; Confidential Data File 1; controlled access via GBIF upon publication. Adding both colonies to our range by assuming that they are either no longer surviving (minimum estimate) or not larger than they were in the 1990s (maximum estimate) we obtain a total for Mauritius of 3525–3577 nests.
On Réunion, our systematic counting including the seven largest colonies yielded a total of 13,669 nests (range: 13,346 to 14,204 nests). In contrast to the Mauritius case, there do not exist a series of earlier colony estimates to make informed inferences of temporal change on Réunion. Based on our random sampling and applying the adapted formula of Bibby et al. (2000) we estimate the number of nests in Réunion colonies not systematically counted to be 1782 (confidence interval: 510–3714 nests; Methods S1). Combining our systematic count with our estimate of the number of nests not systematically counted, we obtain a total for Réunion of 15,451 nests (range: 13,856– 17,918 nests).
Population abundance
Our nest counts yield maximum and minimum estimates for the number of active nests on both Mauritius and Réunion. Based on our calculations (Eq. 1), and rounding all figures to the nearest hundred, we provide the first quantitatively based estimate of the population size of A. f. saffordi on the island of Réunion: 46,500 individuals (range: 39,600–53,500). For Mauritius, we estimate the population size of A. f. francicus as 10,400 mature individuals (range: 10,100–10,700). In total, this yields a population estimate of 56,900 mature individuals for the two island populations of A. francicus combined, with a lower bound estimate of 49,700 and an upper bound estimate of 64,200 mature individuals.
Conservation status assessment
Our data supports a change in the conservation status of A. francicus from Near Threatened to Threatened (Endangered; B2, b(i,ii,iii,iv,v) + c(i,ii,iii,iv)) based on the IUCN red list guidelines (IUCN 2022). We calculated the extent of occurrence (EOO) as 12,371 km2 and the area of occupancy as 132 km2 (Figure 2), which fall under the classification of Vulnerable under Criterion B1 and Endangered under B2 respectively. The species should be listed under the highest threat category for which the taxon qualifies, in this case Endangered per criterion B2. As per the guidelines, we furthermore expect (b) a continuing decline in (i) EOO, (ii) AOO, (iii) quality of habitat, (iv) number of locations or subpopulations, (v) number of mature individuals, as well as, (c) extreme fluctuations in (i) EOO, (ii) AOO, (iii) number of locations or subpopulations, and (iv) number of mature individuals. Furthermore, our findings show that the population of A. francicus on Mauritius, now consists of only slightly more than 10,000 mature individuals (c. 10,400 individuals). Our projection of the decline and extreme fluctuations in area and quality of habitat, number of locations and mature individuals are due to the numerous conservation threats facing the Mascarene Swiftlet (see Discussion).
DISCUSSION
Given significant anthropogenic threats to Mascarene endemic vertebrates and the current economic wealth of one of the two islands in the archipelago (Réunion being a French overseas department), one might imagine that the limited remaining native vertebrate fauna that have so far survived anthropogenic activities would be both well-known, and well protected. Our results for Mascarene Aerodramus provide a good example in which we can question both of these expectations. First, our genetic data show that the two Mascarene island populations exhibit 0.8% genetic divergence in the mitochondrial gene ND2, which is in accordance with Kirwan et al.'s (2018) recognition of two cryptic taxa, at least meriting subspecific status. Second, our population size estimates based on count data for the two islands are the first such estimates for Mascarene Aerodramus overall, including an estimate for Réunion for the first time. Third, even conservatively refraining from splitting the two island populations as species (both of which would then warrant classifying as IUCN Endangered), we find that the existing species, Aerodramus francicus, warrants raising to IUCN Endangered status. Fourth, we find cause for concern for both island populations, with the Mauritius swiftlet population being the more extreme case, both in terms of current absolute population size, and the declines we document for particular colonies which were previously surveyed in the 1990s (Middleton 1998b, 2000).
Phylogeny and genetic status of Mascarene Aerodramus
Our ML and Bayesian analyses of the mitochondrial ND2 gene (Figure 1, S1) demonstrate the existence of two lineages within the Mascarene Swiftlet that exhibit 0.8% HKY pairwise divergence (Table 1), each endemic to a single island (Mauritius and Réunion). Furthermore, our BEAST2 analysis calibrated using externally calculated rates of substitution (Lerner et al. 2011) supports their divergence at approximately 0.1 million years ago. Our concatenated ML and Bayesian analyses (Figure S2, S3) are largely congruent with the ND2 results when support values are taken into account. However, the Mauritian population is not recovered as monophyletic in these analyses, whereas the Réunion population was monophyletic as is A. francicus as a whole, across the two islands. Given that the single gene maximum likelihood analysis of Fib7 demonstrated that the nuclear gene provides little resolution (only four out of 30 nodes showed BS > 75), it seems likely that resolution in the concatenated analyses is adversely affected by the inclusion of Fib7 gene. Therefore, the Fib7 gene is largely uninformative in phylogenetic analysis which is likely due to incomplete lineage sorting (ILS) on the timescale involved.
Species delimitation
The concomitant use of ASAP and BPP, employing barcode gap and coalescent approaches, respectively, allows us to compensate for the different limitations of each of the methods. Indeed, ASAP does not require any starting assumption or hypothesis concerning the number of species nor their cladistic organisation, being based on the barcode gap principle and therefore on the calculation of genetic distances (Puillandre et al. 2021). On the other hand, the user is expected to select the substitution model (JC69, K80 or Simple Distance), and the analysis can only be carried out on one gene at a time (Puillandre et al. 2021). Conversely, the operation of BPP relies on the use of a predetermined guide topology to delimit species, and it is necessary to assign individuals to hypothetical species a priori (Flouri et al. 2018). It should be noted that BPP heuristically allows the use of topology and branch length uncertainties for multiple loci (Flouri et al. 2018). The posterior probability values supporting the recognition of Mascarene Swiftlet lineages of Mauritius and Réunion as separate species are high (PP = 1.0), even higher than those of the other previously recognised species (P > 0.97). Therefore, for Mascarene Aerodramus, results from both ASAP and BPP methods are consistent in delimiting the Mauritius and Réunion lineages as separate species. The further benefit of using species delimitation methods is that they are replicable. As such, they constitute a comparable and objective approach to taxonomy (Fujita et al. 2012).
Taxonomy
Our analyses concord well with the classification proposed by Kirwan et al.'s (2018) that the two island populations should be considered separate taxa. Namely, the single-island endemic subspecies Aerodramus francicus francicus (J.F. Gmelin 1789) from Mauritus and Aerodramus francicus saffordi (Kirwan et al. 2018) from Réunion. Our analysis provide an indication that these lineages are genetically diverged, potentially justifying a revision of species status. However, since our nuclear marker shows incomplete lineage sorting (ILS) and a lack of resolution, we prefer to be conservative and avoid splitting at this stage. Nonetheless, with a view to a future split when more supporting data is obtained, we present here evidence for the recognition of two species under the Phylogenetic (PSC) and Morphological Species Concepts, while acknowledging the need for extra data (ideally extra nuclear genes, or clear evidence from behavioural differences) to support the hypothesis that the Mascarene Swiftlet comprises two species.
We discuss five lines of evidence that offer support for the recognition of two species. First, we demonstrate genetic divergence between A. f. saffordi and A. f. francicus of 0.8% in the mitochondrial ND2 gene and 1% in the Cytb gene. This level of divergence in these genes is in the same range (and sometimes in excess) of that found between other pre-established species within the genus Aerodramus. Second, employing independent genetic loci (ND2 and Fib7) our MSC models implemented in the BPP program consistently inferred separate species for Réunion and Mauritius with high support (PP = 1.0), even higher than those of the other previously recognised species (P > 0.97). Third, the documented widespread existence of a barcode gap in genetic divergence between interspecies variation and intraspecies variation (e.g. Meyer & Paulay 2005, Puillandre et al. 2012) is consistent with such a gap meeting the criterion of diagnosability under the PSC (De Queiroz 2007, Sangster 2014). The barcode gap is a central foundation of the ASAP method we implemented for our dataset. This method places sequences into groups and assigns those groups a ranked ASAP score based on the barcode gap width (Puillandre et al. 2021). For ND2, the gene in which such a barcode gap was identified for all pre-established species, all highest-ranking partitions grouped A. f. saffordi and A. f. francicus as separate species, regardless of the distance metric used. The mitochondrial barcode gap is widely applied and regarded as an effective method within avian taxonomy, and for most cases sister taxa can be easily identified using single mtDNA gene barcode gaps (Baker et al. 2009, Hill 2016).
Fourth, based on mitochondrial data from 16 Mascarene Swiftlets (eight from each island), we have shown using phylogenetic Bayesian inference and maximum likelihood analysis that A. f. francicus and A. f. saffordi are reciprocally monophyletic with strong support (the two lineages are estimated to have diverged 0.1 million years ago). Such reciprocal monophyly conforms with the existence of two species under a phylogenetic species concept in which each member of any single species has a shared and unique evolutionary history, being descended from a common ancestor and possessing defining derived traits. Lastly, based on current norms in taxonomy, it is usually considered ideal that evidence from multiple sources is implemented when describing new species (Schlick-Steiner et al. 2009). As such, previously described morphological differences described by Kirwan et al. (2018) between A. f. saffordi and A. f. francicus could support a split under the Morphological Species Concept. Both field and preliminary museum series descriptions pointed to visible differences in plumage. A subsequent closer investigation found consistent differences in plumage colouration including quantitative differences in the width of the white rump patch (Kirwan et al. 2018). Further biometric analyses showed differences in the length of the tail fork (Kirwan et al. 2018). That these differences are geographically discrete is consistent with the existence of two species, especially given that Aerodramus swiftlets are commonly known for lacking distinguishable morphological characteristics (Chantler et al. 2000).
Figure 3.
Examples of human disturbance at breeding locations of A. francicus on Mauritius. (A) Rubbish dump almost blocking the entrance of a cave, (B) religious ceremony being held inside nesting cave, (C) Domestic Cat lurking in swiftlet flyway, (D) burnt car tyres in nesting cave (alongside pile of rubbish). Photos by Y.A.

Although these phylogenetic analyses were performed utilising both nuclear and mitochondrial data from multiple Aerodramus species, it is reasonable to apply caution in their interpretation. The interpretation of the phylogenetic results relies heavily on a single mitochondrial gene (ND2). Moreover, the sole nuclear gene employed (Fib7) does not provide adequate information due to low resolution and the effects of incomplete lineage sorting (ILS) within the genus Aerodramus. This lack of evidence from different genetic sources is our primary rationale behind the decision to refrain from a taxonomic revision splitting A. francicus into two species, pending further data. One possible explanation for the divergence we observe between island populations in mitochondrial data is a reduction in gene flow between them. However, without more data from the nuclear genome, we cannot reliably assess how much gene flow actually occurs. Consequently, in addition to the potential to add vocal or behavioural data, phylogenetic and species delimitation analyses should include a larger number of independent loci, ideally on a genome-wide scale, in order to reach a conclusion on the species status of the swiftlet taxa of Mauritius and Réunion.
Population size estimates, threats and conservation needs
Our estimation of 7100 breeding individuals on Mauritius is larger than previous estimates based on Middleton's (1998b, 2000) counts of 2244–2610 breeding individuals. This difference can be at least partly accounted for by an increase in numbers in two colonies (names withheld), one of which represents a 10-fold increase since the 1990s, to 1387 nests today. Another reason for the difference could be that a more thorough survey (as performed in this study) logically yields higher numbers. However, there are strong signs that these isolated points of brighter news for the conservation status of the swiftlet on Mauritius should not be interpreted as evidence for a secure status of this island population overall. Rather, the loss of the two largest colonies documented by Middleton (1998b, 2000) for the 1990s, and the current deteriorating status of the caves that once held them (Hammond et al. 2015) is a significant cause for concern. Surinam Cave, that held the largest colony in the 1990s estimated by Middleton (1998b, 2000) as 600–700 swiftlets, has since suffered not only from the construction of a building over one entrance, but is also used for illegal rubbish dumping that entirely blocks the other entrance. Similarly, Palma Cave, the second largest in the 1990s, had much construction rubble and rubbish dumped in it, until it was recently cleaned by members of a local NGO (Ecosystem Restoration Alliance Indian Ocean). In both cases, no swiftlets survived or remained. In January 2020, we found (and removed) poles used for poaching that had not been present a few months earlier at a large colony (name withheld for conservation security). Furthermore, at both a large colony and a smaller one we found the remains of burnt tyres.
Beyond the specifics of threats incurred at key Mauritian colonies, the main threats facing A. francicus across its range (Mauritius and Réunion) are the degradation, disturbance and destruction of nesting-caves, and various introduced predators (mammals, reptiles, birds and invertebrates). The Domestic Cat Felis catus has been observed in caves hosting bats and swiftlets (Figure 3C). Bird predation by Domestic Cats is a serious and often neglected worldwide conservation threat (Trouwborst et al. 2020). In the Mascarenes, it has been observed that cats, which are an alien species in the archipelago, can leap and catch flying swiftlets (D. Strasberg pers. comm.). Similar observations of predation by cats were made multiple times on other island swiftlets (e.g. Manchi & Sankaran 2009), and Tidemann et al. (1994) recorded swiftlets in the diet of introduced cats on Christmas Island. Invasive cockroaches have also been observed around swiftlet nests in the Mascarenes. Cockroaches are known to reduce the breeding success of Edible-nest Swiftlet Aerodramus fuciphagus by feeding on their saliva-built nests (Manchi & Sankaran 2009). Therefore, they are also likely to negatively affect A. francicus. Alien crows, rats, snakes and ants also occur in the Mascarenes and are common predators of adult birds, newly hatched chicks or fledglings.
Rapid population growth and urban expansion have increased pressure on Mascarene wildlife and natural areas, especially in Mauritius (Norder et al. 2017). A recent assessment on environmentally sensitive areas (ESA) in Mauritius has found an unusually high proportion of caves within and close to urban areas (Hammond et al. 2015). Unfortunately, the largest remaining colonies of the Mauritian swiftlet also disproportionally involve caves that are in close proximity of urban areas and are at risk of urban expansion (Hammond et al. 2015, observations by BHW). Caves previously inhabited by swiftlet colonies have already been observed to have their entrances completely blocked after dumping of rubbish (Middleton 1998a,b). It is also common for people to burn tyres inside caves (Cheke & Hume 2008). Indeed, remains of burnt tyres were observed during recent nest surveys. The toxic fumes from these fires are likely to have devastating consequences to all cave-living creatures. Furthermore, one striking example of urbanisation on Mauritius interfering with the breeding habitat of the Mauritius swiftlet is our discovery of 245 nests in a disused building near Quatre Bornes. Sadly, not long after this discovery, the empty building was converted into a commercial centre and currently no swiftlets remain. Therefore, we did not include this former breeding location in our calculations of population size, nor EOO and AOO. It is worth noting that we can assume that this particular colony (consisting of approximately 715 swiftlets) was entirely eradicated, hence resulting in a substantial recent population decline. Another threat is the illegal harvesting of the saliva-built nests of the swiftlet for bird's nest soup (Safford & Hawkins 2013). The decline of the Mauritius swiftlet population over the past years can be partly attributed to illegal harvesting. Our nest surveys revealed signs that poaching is currently ongoing.
Although the larger population size of A. f. saffordi and more mountainous topography, and greater native forest of Réunion might provide some buffering, Réunion's A. f. saffordi faces similar threats to its existence to those of A. f. francicus of Mauritius. Like A. f. francicus, large fluctuations in population size of A. f. saffordi are expected due to anthropogenic activities and stochastic weather events such as extreme variability in rainfall (Réchou et al. 2019) and the increasing intensity of cyclones due to climate change (Vidya et al. 2021). Poaching likely remains a significant threat island-wide, significant signs of which were observed at Trou Z'armand, Éperon (Saint-Gilles) in 2019 (observations by BHW and JMP). Furthermore, contrary to the case in Mauritius, many other colony-bearing caves are located within Réunion National Park (established in 2007), which so far usually accords them protection in theory but not in practice. A notable example is the fourth largest Réunion colony located in the natural arch-like cave of La Chapelle, in the Cirque de Cilaos. Significant traffic of pedestrian sightseers through the cave continually disturbs the nesting swiftlets causing chicks to fledge prematurely and fall to their death.
Given our assessment of the threatened status of A. francicus there is an urgent need for appropriate protection and restoration of the main swiftlet nesting sites of both Mauritius and Réunion. Currently there is no legislation in place to protect the nesting sites of A. f. francicus. This gives a bleak prospect for the Mauritius taxon in terms of the likelihood of population decline and the probability of extinction, particularly given the ongoing and increasing threats. Awareness that these caves harbour the natural heritage of Mauritius should be actively promoted among local communities and tourism agencies. Protection of these caves would not only benefit colonies of the Mauritius swiftlet, but also protect many species that share this habitat, such as the endemic Natal Free-tailed Bat Mormopterus acetabulosus (Goodman et al. 2008) and other cave-associated biodiversity, including a subspecies of louse Dennyus carljonesi forresteri uniquely hosted by A. f. francicus of Mauritius (Clayton et al. 1996). A lack of public awareness of the importance of caves for swiftlets and Mascarene natural heritage is a problem that needs addressing on both Mauritius and Réunion. However, prospects for A. f. francicus are particularly bleak, not only because the threats weigh on a swiftlet population that is four times smaller than the population of A. f. saffordi, but also because none of the caves nor colonies they hold are currently protected in practice. Mauritian caves have been classified as ‘Ecologically Sensitive Areas’ (ESA) needing protection (Hammond et al. 2015) and included as such in the ESA Bill drafted in 2009. Unfortunately, the Bill was never enacted and there currently exists no serious indication that it will be enacted.
Therefore, we recommend considering signage to explain the environmental perils of disturbance to sightseers, as well as possible park guard attendance to regulate access in peak visitor periods on both Mauritius as well as Réunion. We note that such measures to improve the protection of caves could be beneficial to other cave-dwelling species such as the endemic bats, M. acetabulosus on Mauritius and M. francoismoutoui on Réunion (Goodman et al. 2008). Furthermore, we recommend immediate measures to prevent swiftlets from using unsuitable human infrastructure as breeding sites. The swiftlets tend to colonize anthropogenic infrastructure such as tunnels, bridges and disused buildings, where they are exposed to regular disturbance or more extreme harm from human activities. It is therefore recommended to block access to certain anthropogenic infrastructures so that new colonies can no longer be established. In cases in which a colony has already established, we recommend closing pedestrian access while ensuring safe passage for swiftlets.
Conclusion
Detailed information on the taxonomic and conservation status of species worldwide is key in addressing the ongoing biodiversity crisis. At a global scale, birds are among the higher taxa that have been longest and most-intensively studied, being both readily distinguishable and appealing to humans. Interest in Mascarene birds relate not only to these global-scale factors, but also to the fact that the modern avifauna, with just 20 native terrestrial breeding species (including Aerodramus), is but a moderate portion of a documented avifauna that was once much more diverse. As in many volcanic archipelagos worldwide, birds are one among relatively few taxa (including mammals, reptiles and land snails) for which we have significant information on the biodiversity that has been lost due to extinction since human colonization in the 1600s (1638 for Mauritius and 1665 for Réunion). In the case of birds, this relates both to the preservation of hard parts in the form of subfossils, particularly under conducive preservation conditions (Rijsdijk et al. 2009), as well as to their appeal to the first humans arriving in a pristine archipelago 400 years ago, who documented their observations in paintings and informal reports (reviewed by Cheke & Hume 2008).
Our results counter what might intuitively be assumed about such an avifauna, that it would be both well-studied and well-protected today, especially considering that the Mascarene archipelago currently benefits from some of the highest levels of economic development in the Indian Ocean region and (in the case of Réunion) in the European Union. In combination with data from an earlier study (Kirwan et al. 2018), our results provide support for the existence of two Aerodramus species in the Mascarene archipelago. Given this finding, we suggest that it would be prudent that conservation decision makers take action for the two island taxa as though they were separate species. However, we currently refrain from splitting into two species due to a lack of genome-wide data which would provide more clarity on the level of gene flow and degree of reproductive isolation. Obtaining such data may likely result in a species split in the near future. Each of the two resulting species would then be classified as Threatened (Endangered) based on our data and conservation assessment, employing IUCN criteria. Nonetheless, even without this split, our data and conservation assessment support an update in threat status for A. francicus from Near Threatened to Threatened (Endangered) under IUCN criteria. Particularities of the genus Aerodramus go some way towards explaining this threat status and may explain why other Aerodramus species are currently listed by the IUCN as Threatened (three out of 23), these also being endemic to oceanic islands (Atiu, Mariana, Seychelles). The flying abilities of Aerodramus swiftlets has led them to disperse all over the world. Furthermore, they form colonies with a high-fidelity to their nesting caves, for which there is evidence that micro-climatic requirements are unusually specific (Nguyen Quang et al. 2002). Their natal philopatry limits their dispersal and gene flow to nearby islands after first colonisation, which has likely facilitated high levels of speciation in the genus (Rheindt et al. 2014). Their unique nesting requirements also render them particularly vulnerable to habitat degradation or destruction. Nonetheless, the situation of Mascarene Aerodramus as being both threatened and relatively neglected is not an isolated one among recent studies of Mascarene endemic vertebrates (Probst & Deso 2001, Florens & Baider 2019, Bhanda & Florens 2021, Bunsy & Florens 2021, Seegobin et al. 2022). Greater environmental conscience is needed in the Mascarenes to curb the loss of its unique biodiversity.
ACKNOWLEDGEMENTS
For funding, we thank Electricité de France and the French Agence Nationale de la Recherche (ANR) under grant ANR-20-CE02-0009. We also thank Louise Breton for performing many of the DNA extractions and PCR amplifications. Furthermore, we are thankful to G.J. Middleton for his first effort to estimate the population size of the swiftlet on Mauritius, for extensively sharing his knowledge on the locations of breeding habitats, and for his comments on the manuscript. We also thank SEOR volunteers and multiple SEOR staff including Damien Chiron, Francois-Xavier Couzi and Nicolas Laurent for help with obtaining samples from the SEOR centre de soins, for participation in some of the colony surveys and for help in accessing SEOR documents. We further thank Roger Safford for help in accessing Mascarene Swiftlet literature, and for discussing how this has been interpreted.
REFERENCES
Appendices
SAMENVATTING
De eilandengroep de Mascarenen in de Indische Oceaan is ongeveer 400 jaar geleden door mensen gekoloniseerd. Sindsdien heeft er een extreme degradatie van ecosystemen plaatsgevonden en zijn talloze soorten uitgestorven. De Mauritiussalangaan Aerodramus francicus, een van de overlevende soorten van de eilandengroep, bouwt zijn nesten in de grotten van Mauritius en Réunion. De salanganen van het geslacht Aerodramus zijn moeilijk op naam te brengen, doordat zij weinig duidelijke morfologische kenmerken hebben. Ze zijn bovendien bijzonder kwetsbaar voor verstoring door de mens. Wij hebben in 2019–2021 onderzocht of de twee eilandpopulaties genetisch van elkaar verschillen om inzicht te krijgen in hun taxonomische status. De twee eilandpopulaties bleken genetisch 0,8% van elkaar te verschillen en op grond van het mitochondriale ND2-gen uit dezelfde voorouder te stammen. De evolutionaire divergentie binnen het gehele genus Aerodramus kon op grond van de verkregen gegevens niet worden opgelost. We kunnen daardoor nog niet zeggen of de twee populaties verschillende soorten zijn. Op grond van onze tellingen schatten wij dat er 10.100–10.700 volwassen individuen op Mauritius voorkomen en 39.600–53.500 op Réunion. Op grond van de geringe populatiegrootte, het kleine leefgebied, de optredende habitatdegradatie en menselijke verstoring rechtvaardigen dat de status van de vogels op Mauritius en Réunion volgens de IUCN-richtlijnen worden bijgesteld naar ‘Bedreigd’. Bescherming en verbetering van het leefgebied van de soort op beide eilanden is cruciaal om te voorkomen dat ook deze endemische soort zal uitsterven.
Supplementary Material is available online www.ardeajournal.nl/supplement/s112-###-###.pdf
SUPPLEMENTARY MATERIAL
Methods S1. Formula and figures used to estimate the number of swiftlet nests in colonies on Réunion that were not systematically counted.
Following Bibby et al. (2000), an estimate of the number of nests in colonies on Réunion that were not systematically counted was estimated using:
N = nA/a
Where
N = the estimation of total number of nests not systematically counted
A = the total number of colonies that were not systematically counted
a = the number of non-systematically counted colonies drawn at random
n = the number of nests counted in non-systematically sampled colonies drawn at random
Applying the data on colonies not systematically counted, and our random counts:
A = 27
a = 5
n = 330
Therefore, N = 1782.
The confidence limits on this estimate of the number of nests in colonies on Reunion that were not systematically counted are calculated as follows:
Upper limit = n + (mean + 1.96 × SE) × (A–a)
Lower limit = n – (mean + 1.96 × SE) × (A–a)
Therefore, our confidence limits for the estimate of the number of nests in colonies on Réunion that were not systematically counted is 510–3714 nests.
Figure S1.
Bayesian consensus tree inferred for Aerodramus species and outgroups (Apus, Collocalia) from BEAST analysis of the mitochondrial ND2 gene matrix under the TrN+I substitution model. Numbers in bold indicate posterior probabilities (PP) of the corresponding clades and numbers between brackets indicate the node ages in millions of years.

Figure S2.
Maximum likelihood analysis for Aerodramus species and outgroups (Apus, Collocalia) of the concatenated ND2-Fib7 gene matrix based on the GTR substitution model. Bootstrap support values (1000 replicates) are indicated on nodes.

Figure S3.
Bayesian consensus tree for Aerodramus species and outgroups (Apus, Collocalia) inferred from BEAST analysis of the concatenated ND2-Fib7 gene matrix under the TrN+I and GTR substitution models (respectively). Numbers in bold indicate posterior probabilities (PP) of the corresponding clades.